Gene expression regulatory programs
Understanding which regulators of gene expression are crucial for establishing and maintaining cell identity can provide means to correct aberrant cells.
Although the genetic material is identical in all cells of our body, the way in which genes are ‘read’ and ‘expressed’ differs, leading to a wide variety of cell shapes, sizes and functions.
RNA processing is linked to cell fate
Gene expression is a complex process, starting with changes in the chromatin state, going through the synthesis and processing of precursor messenger RNAs (pre-mRNAs), export to the cytoplasm, translation into protein and final decay. As the use of high-throughput sequencing technologies increased, a wealth of data has become available in public repositories. By applying our computational models to the analysis of large data sets, we have uncovered global regulators that affect cellular behavior by acting at distinct steps of the gene expression cascade, particularly in the splicing of pre-mRNAs, the cleavage and addition of a poly adenine tail (polydenylation) as well as in mRNA translation.
High-throughput data combined with computational models
For the majority of human genes, pre-mRNAs can be processed in different ways, leading to multiple transcript ‘isoforms’ per gene. By analyzing many mRNA expression data sets, obtained from organisms ranging from mice to humans, we have identified evolutionarily conserved regulators of the splicing – the process by which pre-mRNAs are cut and pasted to make the mRNAs - that are key to the establishment of specific cell types.
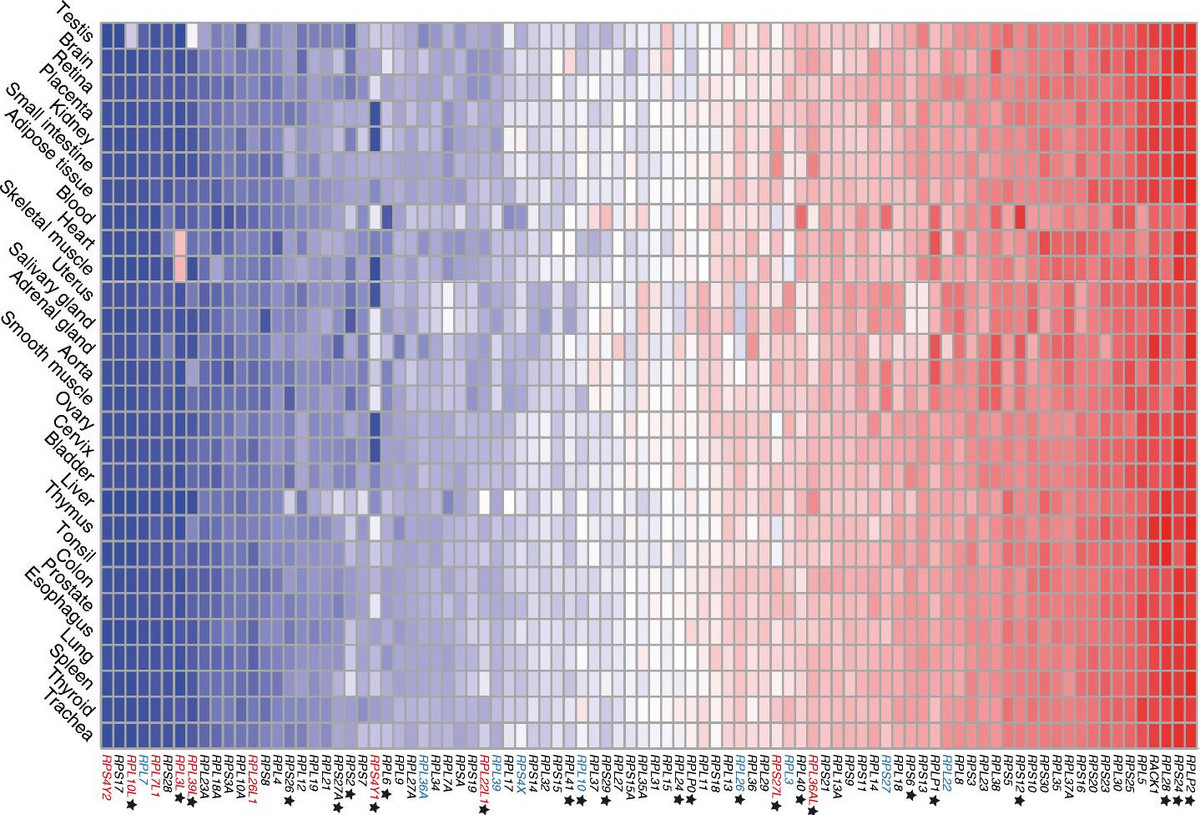
The length of mRNAs is also linked to cells’ state, highly proliferative cells such as activated lymphocytes and cancer cells expressing short transcripts, whereas neurons express long transcripts. A change in mRNA length implies a change in the mRNA’s interactions, and thus alternative polyadenylation has far reaching implications. We have uncovered two regulators of transcript length, the cleavage factor I, and the heterogeneous ribonucleoprotein C. Currently, we study polyadenylation in the context of cancer. Surprisingly, we also found that the expression patterns of components of the mRNA translation machinery distinguish between normal and cancer cells.
Towards new strategies for manipulating cell fate
Ultimately, we hope that by manipulating expression of factors that define specific cell states we can change cell fates in a highly controlled manner.